Data Frames, Basic Indexing, Reading/Writing Data
Overview
Class Date: 9/4/2025 -- In Class
Teaching: 90 min
Exercises: 30 minQuestions
What is a data frame?
How does indexing differ for data frames relative to basic data structures?
How do I access data frame subsets?
How do I read data from a .csv or .txt file into R?
How do I write data to a .csv or .txt file?
Objectives
Understand the structure and properties of data frames in R.
Select individual values and subsections from data.
Perform operations on a data frame.
Understand the three primary ways R can access data inside a data frame.
Be able to apply the basic functions for reading and writing tabular data stored in common text file formats (.txt, .csv).
Understand the key arguments for importing data properly, such as
headerandstringsAsFactors.Understand the key arguments for exporting the data properly, such as
row.names,col.names, andquote.Combine different methods for accessing data with the assignment operator to update subsets of data.
In Class
Data Frames
The data frame is the most important data type in R. It is the de facto data structure for most tabular data and what we will use for calculating statistics and running other analyses. Data frames have some similarities to matrices. For example, both are two-dimensional structures and can be similarly indexed using []. However, data frames have several features that distinguish them from matrices and make them more generally useful to store and manipulate datasets.
Data frames have the following characteristics:
- Each column must contain a single data type
- Different columns can have different data types
- All columns must have the same number of elements (i.e. rows)
- Each column has a names can be directly called using the
$operator - Like matrices, data frames can be indexed using the
df[row,column]notation.
As with other data types and structure, data frames can be generated manually using the data.frame() function:
dat <- data.frame(id = letters[1:10], x = 1:10, y = 11:20)
dat
id x y
1 a 1 11
2 b 2 12
3 c 3 13
4 d 4 14
5 e 5 15
6 f 6 16
7 g 7 17
8 h 8 18
9 i 9 19
10 j 10 20
Note the structural difference when compared to the same object coerced to a matrix, particularly the different data types in each column:
m <- as.matrix(dat)
m
id x y
[1,] "a" " 1" "11"
[2,] "b" " 2" "12"
[3,] "c" " 3" "13"
[4,] "d" " 4" "14"
[5,] "e" " 5" "15"
[6,] "f" " 6" "16"
[7,] "g" " 7" "17"
[8,] "h" " 8" "18"
[9,] "i" " 9" "19"
[10,] "j" "10" "20"
Converting a data frame to a matrix enforces the “single data type” requirement of the latter by coercing all elements to the most general data type already present (in this case character). If we try to reverse the conversion (using the as.data.frame() function) we lose the original data structure:
m.dat <- as.data.frame(m)
m.dat
id x y
1 a 1 11
2 b 2 12
3 c 3 13
4 d 4 14
5 e 5 15
6 f 6 16
7 g 7 17
8 h 8 18
9 i 9 19
10 j 10 20
The attributes are also distinct for the two data types.
class(m)
[1] "matrix" "array"
class(dat)
[1] "data.frame"
attributes(m)
$dim
[1] 10 3
$dimnames
$dimnames[[1]]
NULL
$dimnames[[2]]
[1] "id" "x" "y"
attributes(dat)
$names
[1] "id" "x" "y"
$class
[1] "data.frame"
$row.names
[1] 1 2 3 4 5 6 7 8 9 10
dim(m)
[1] 10 3
dim(dat)
[1] 10 3
The above features mean that matrices and data frames are suitable for different data types. For instance, matrices are optimized for data of the same type, and particularly data with a inherently two-dimensional structure–e.g. image data stored as pixel intensities at different x-y coordinates. Data frames are optimized for data sets with mixed data types–e.g. patient data with multiple phenotypes, such as patient ID (character), sex (character), body weight (numeric), and smoking status (logical). The mixed structure of data in data frames makes them non-suitable to matrix operations (e.g. matrix multiplication, transposition).
Here is some additional information on data frames:
- A data frame is created by the
read.csv()andread.table()when importing the data into R (see below). - Assuming all columns in a data frame are of same type, data frame can be converted to a matrix with
data.matrix()(preferred) oras.matrix(). Otherwise type coercion will be enforced and the results may not always be what you expect. - Data frames can be created de novo with the
data.frame()oras.data.frame()functions. - Like matrices, the number of rows and columns can be queried with
nrow(dat)andncol(dat), respectively. - Data frames can have additional attributes such as
rownames(), which can be useful for annotating data, likesubject_idorsample_id. - Row names are often automatically generated to reflect the row number:
1, 2, ..., n. Consistency in numbering of row names may not be honored when rows are reshuffled or when a data frame is subsetted. - A common way to use data frames is with columns as “variables” (e.g. body weight, blood pressure) and rows as “patients” or “observations” (e.g. “subject 1”, “subject 2”, …).
Just as a matrix in R is a specialized vector, a data frame is a specialized list. More on this in today’s On Your Own section. For now, the following table summarizes the one-dimensional and two-dimensional data structures in R in relation to diversity of data types they can contain.
| Dimensions | Homogeneous | Heterogeneous |
|---|---|---|
| 1-D | atomic vector | list |
| 2-D | matrix | data frame |
Sample data in R
R includes a number of built-in data sets that can be used to examine and test different functions and operators. The variable iris contains a pre-defined data frame that is automatically loaded into memory when R is initialized. We will use iris to begin learning how to interact with data frames.
Let’s start by looking at the documentation for the iris data set:
?iris
For reference, here are images of the flowers from the three iris species from the iris dataset:

There are many ways to interact with data frames and get information about their contents. We have seen some of these previously:
Useful Data Frame Functions
head()- shows first 6 rowstail()- shows last 6 rowsdim()- returns the dimensions of data frame (i.e., number of rows and number of columns)nrow()- number of rowsncol()- number of columnsstr()- structure of data frame - name, type and preview of data in each columnnames()orcolnames()- both show thenamesattribute for a data framesapply(dataframe, class)- shows the class of each column in the data frame
I find head(), which displays the first 6 rows and all columns of the data frame, particularly useful to get a feel for the contents and organization of a new data frame. Let’s take a quick look at iris with head() and dim():
head(iris)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
head(iris, 2) # you can look at more or fewer rows if 6 is not what you want
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
dim(iris)
[1] 150 5
We know that data frames can store different data types in different columns, but how do we tell which data types are present in a specific data frame?
class(iris)
[1] "data.frame"
mode(iris)
[1] "list"
typeof(iris)
[1] "list"
The usual functions for looking at the data type of basic variables and data structures don’t seem to work. This is where the str() is particularly useful:
str(iris)
'data.frame': 150 obs. of 5 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
str() gives a lot of useful information about data frames:
- number of rows (i.e.
150 obs.) - number of columns (i.e.
5 variables) - the name of each column (e.g.
$Sepal.Length) - the data type of each column (e.g.
num) - the first 10 entries in each column (e.g.
5.1, 4.9, 4.7, ...)
This is the same information displayed if you toggle the arrow for a data frame variable in the Environment panel (by default this is in the upper-right panel of RStudio), for instance the dat variable we created earlier. Clicking on a variable opens a basic spreadsheet representation of the table in the Source Editor panel (upper-left by default).
Indexing data frames
We examined basic indexing using vectors and matrices in last week’s lessons. These work the same for data frames. There are really three primary ways for accessing specific data inside data frames:
- By index
- By name
- By logical vector
These can be combined to rapidly extract desired data subsets within data frames. You will be exploring these in depth On Your Own, but here I will give you the flavor of each.
By index
Because data frames are rectangular, elements of data frame can be referenced by specifying the row and the column index in single square brackets (similar to matrices).
iris[1, 3]
[1] 1.4
Like matrices, we can also ask for multiple columns and rows using the : operator:
iris[1:20, 2:3]
Sepal.Width Petal.Length
1 3.5 1.4
2 3.0 1.4
3 3.2 1.3
4 3.1 1.5
5 3.6 1.4
6 3.9 1.7
7 3.4 1.4
8 3.4 1.5
9 2.9 1.4
10 3.1 1.5
11 3.7 1.5
12 3.4 1.6
13 3.0 1.4
14 3.0 1.1
15 4.0 1.2
16 4.4 1.5
17 3.9 1.3
18 3.5 1.4
19 3.8 1.7
20 3.8 1.5
… or for non-contiguous subsets using lists of indices:
iris[c(1:10,20:25), c(1,3)] # note the use of sequences within the `c()` list
Sepal.Length Petal.Length
1 5.1 1.4
2 4.9 1.4
3 4.7 1.3
4 4.6 1.5
5 5.0 1.4
6 5.4 1.7
7 4.6 1.4
8 5.0 1.5
9 4.4 1.4
10 4.9 1.5
20 5.1 1.5
21 5.4 1.7
22 5.1 1.5
23 4.6 1.0
24 5.1 1.7
25 4.8 1.9
As with other data types, leaving a dimension blank is interpreted as return all values in that dimension:
iris[2,] # return row 2
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
2 4.9 3 1.4 0.2 setosa
iris[,2] # return column 2
[1] 3.5 3.0 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 3.7 3.4 3.0 3.0 4.0 4.4 3.9 3.5
[19] 3.8 3.8 3.4 3.7 3.6 3.3 3.4 3.0 3.4 3.5 3.4 3.2 3.1 3.4 4.1 4.2 3.1 3.2
[37] 3.5 3.6 3.0 3.4 3.5 2.3 3.2 3.5 3.8 3.0 3.8 3.2 3.7 3.3 3.2 3.2 3.1 2.3
[55] 2.8 2.8 3.3 2.4 2.9 2.7 2.0 3.0 2.2 2.9 2.9 3.1 3.0 2.7 2.2 2.5 3.2 2.8
[73] 2.5 2.8 2.9 3.0 2.8 3.0 2.9 2.6 2.4 2.4 2.7 2.7 3.0 3.4 3.1 2.3 3.0 2.5
[91] 2.6 3.0 2.6 2.3 2.7 3.0 2.9 2.9 2.5 2.8 3.3 2.7 3.0 2.9 3.0 3.0 2.5 2.9
[109] 2.5 3.6 3.2 2.7 3.0 2.5 2.8 3.2 3.0 3.8 2.6 2.2 3.2 2.8 2.8 2.7 3.3 3.2
[127] 2.8 3.0 2.8 3.0 2.8 3.8 2.8 2.8 2.6 3.0 3.4 3.1 3.0 3.1 3.1 3.1 2.7 3.2
[145] 3.3 3.0 2.5 3.0 3.4 3.0
Note that the column version actually returns a vector of the data type in the requested column number, rather than a data frame with a single column:
class(iris[,2])
[1] "numeric"
What if we only ask for one dimension in data frame?
Asking for a single index from a data frame
What do you expect if we query
iriswith just one index?iris[3]Solution
iris[3]Petal.Length 1 1.4 2 1.4 3 1.3 4 1.5 5 1.4 6 1.7 7 1.4 8 1.5 9 1.4 10 1.5 11 1.5 12 1.6 13 1.4 14 1.1 15 1.2 16 1.5 17 1.3 18 1.4 19 1.7 20 1.5 21 1.7 22 1.5 23 1.0 24 1.7 25 1.9 26 1.6 27 1.6 28 1.5 29 1.4 30 1.6 31 1.6 32 1.5 33 1.5 34 1.4 35 1.5 36 1.2 37 1.3 38 1.4 39 1.3 40 1.5 41 1.3 42 1.3 43 1.3 44 1.6 45 1.9 46 1.4 47 1.6 48 1.4 49 1.5 50 1.4 51 4.7 52 4.5 53 4.9 54 4.0 55 4.6 56 4.5 57 4.7 58 3.3 59 4.6 60 3.9 61 3.5 62 4.2 63 4.0 64 4.7 65 3.6 66 4.4 67 4.5 68 4.1 69 4.5 70 3.9 71 4.8 72 4.0 73 4.9 74 4.7 75 4.3 76 4.4 77 4.8 78 5.0 79 4.5 80 3.5 81 3.8 82 3.7 83 3.9 84 5.1 85 4.5 86 4.5 87 4.7 88 4.4 89 4.1 90 4.0 91 4.4 92 4.6 93 4.0 94 3.3 95 4.2 96 4.2 97 4.2 98 4.3 99 3.0 100 4.1 101 6.0 102 5.1 103 5.9 104 5.6 105 5.8 106 6.6 107 4.5 108 6.3 109 5.8 110 6.1 111 5.1 112 5.3 113 5.5 114 5.0 115 5.1 116 5.3 117 5.5 118 6.7 119 6.9 120 5.0 121 5.7 122 4.9 123 6.7 124 4.9 125 5.7 126 6.0 127 4.8 128 4.9 129 5.6 130 5.8 131 6.1 132 6.4 133 5.6 134 5.1 135 5.6 136 6.1 137 5.6 138 5.5 139 4.8 140 5.4 141 5.6 142 5.1 143 5.1 144 5.9 145 5.7 146 5.2 147 5.0 148 5.2 149 5.4 150 5.1Because the data frame is a list at heart, just asking for a single index returns the indicated column.
By name
Data frames (and lists; see On Your Own section) have a special property in that each column is considered a separate element with a unique name that can be used as a handle to call the data in that column. To exploit this feature, you can either use double square brackets ([[]]) or the $:
iris[["Sepal.Length"]]
[1] 5.1 4.9 4.7 4.6 5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1
[19] 5.7 5.1 5.4 5.1 4.6 5.1 4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0
[37] 5.5 4.9 4.4 5.1 5.0 4.5 4.4 5.0 5.1 4.8 5.1 4.6 5.3 5.0 7.0 6.4 6.9 5.5
[55] 6.5 5.7 6.3 4.9 6.6 5.2 5.0 5.9 6.0 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9 6.1
[73] 6.3 6.1 6.4 6.6 6.8 6.7 6.0 5.7 5.5 5.5 5.8 6.0 5.4 6.0 6.7 6.3 5.6 5.5
[91] 5.5 6.1 5.8 5.0 5.6 5.7 5.7 6.2 5.1 5.7 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3
[109] 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6.0 6.9 5.6 7.7 6.3 6.7 7.2
[127] 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6.0 6.9 6.7 6.9 5.8 6.8
[145] 6.7 6.7 6.3 6.5 6.2 5.9
iris$Sepal.Length
[1] 5.1 4.9 4.7 4.6 5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1
[19] 5.7 5.1 5.4 5.1 4.6 5.1 4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0
[37] 5.5 4.9 4.4 5.1 5.0 4.5 4.4 5.0 5.1 4.8 5.1 4.6 5.3 5.0 7.0 6.4 6.9 5.5
[55] 6.5 5.7 6.3 4.9 6.6 5.2 5.0 5.9 6.0 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9 6.1
[73] 6.3 6.1 6.4 6.6 6.8 6.7 6.0 5.7 5.5 5.5 5.8 6.0 5.4 6.0 6.7 6.3 5.6 5.5
[91] 5.5 6.1 5.8 5.0 5.6 5.7 5.7 6.2 5.1 5.7 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3
[109] 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6.0 6.9 5.6 7.7 6.3 6.7 7.2
[127] 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6.0 6.9 6.7 6.9 5.8 6.8
[145] 6.7 6.7 6.3 6.5 6.2 5.9
These named columns can be used in vectorized mathematical operations:
iris$Sepal.Length + iris$Petal.Length
[1] 6.5 6.3 6.0 6.1 6.4 7.1 6.0 6.5 5.8 6.4 6.9 6.4 6.2 5.4 7.0
[16] 7.2 6.7 6.5 7.4 6.6 7.1 6.6 5.6 6.8 6.7 6.6 6.6 6.7 6.6 6.3
[31] 6.4 6.9 6.7 6.9 6.4 6.2 6.8 6.3 5.7 6.6 6.3 5.8 5.7 6.6 7.0
[46] 6.2 6.7 6.0 6.8 6.4 11.7 10.9 11.8 9.5 11.1 10.2 11.0 8.2 11.2 9.1
[61] 8.5 10.1 10.0 10.8 9.2 11.1 10.1 9.9 10.7 9.5 10.7 10.1 11.2 10.8 10.7
[76] 11.0 11.6 11.7 10.5 9.2 9.3 9.2 9.7 11.1 9.9 10.5 11.4 10.7 9.7 9.5
[91] 9.9 10.7 9.8 8.3 9.8 9.9 9.9 10.5 8.1 9.8 12.3 10.9 13.0 11.9 12.3
[106] 14.2 9.4 13.6 12.5 13.3 11.6 11.7 12.3 10.7 10.9 11.7 12.0 14.4 14.6 11.0
[121] 12.6 10.5 14.4 11.2 12.4 13.2 11.0 11.0 12.0 13.0 13.5 14.3 12.0 11.4 11.7
[136] 13.8 11.9 11.9 10.8 12.3 12.3 12.0 10.9 12.7 12.4 11.9 11.3 11.7 11.6 11.0
Default Names
If column names are not specified,
V1, V2, ..., Vnare automatically used as the defaults. For example, if we convert a matrix to a data frame:m <- matrix(1:10, nrow = 2) m.data <- as.data.frame(m) m.dataV1 V2 V3 V4 V5 1 1 3 5 7 9 2 2 4 6 8 10
By logical vector
Recall that a logical vector contains only the special values TRUE and FALSE:
c(TRUE, TRUE, FALSE, FALSE, TRUE)
[1] TRUE TRUE FALSE FALSE TRUE
Data frames and other data structures can accept logical vectors as indexing variables. Usually the length of the vector will be the same as the corresponding dimension of the data frame, with each vector element indicating whether the corresponding row or column in the data frame should be included (TRUE) or excluded (FALSE) in the output data frame. Let’s look at a simple example using dat:
# grab the dimensions of dat
dim(dat)
[1] 10 3
# create a logical vector with 10 elements indicating that
# elements 2, 3, and 7 should be returned
index <- c(F,T,T,F,F,F,T,F,F,F)
# use the created logical vector to index the rows of dat
dat[index,]
id x y
2 b 2 12
3 c 3 13
7 g 7 17
Logical indexing is one of the most powerful subsetting techniques in R. Generally, we won’t be manually defining the index vector, but using R to generate the appropriate logical vector. We will go into more depth in the On Your Own, but let’s look at an example and a few simple exercises to look at how logical indexing may be used in practice.
Looking at the iris dataset, we can use the logical operator == to return the subset of data corresponding to the Iris setosa species. First, let’s examine what the == operator does:
# Which elements of a vector are exactly equal to 1?
x <- c(3,2,1,2,1,2,1)
x == 1
[1] FALSE FALSE TRUE FALSE TRUE FALSE TRUE
The expression x == 1 automatically returns a logical vector of the same length as x with TRUE elements where x is exactly 1 and FALSE elements where x is not exactly 1. Let’s apply this to our iris data set:
# create a vector indicating which elements of the iris "Species"
# variable are equal to "setosa"
i.setosa <- iris$Species == "setosa"
i.setosa
[1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[13] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[37] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[49] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[61] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[73] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[85] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[97] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[109] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[121] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[133] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[145] FALSE FALSE FALSE FALSE FALSE FALSE
# use the vector to extract the subset of the iris data for flowers
# of the "Iris setosa" species
setosa <- iris[i.setosa,] # don't forget to specify "all" columns!
setosa
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5.0 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5.0 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
11 5.4 3.7 1.5 0.2 setosa
12 4.8 3.4 1.6 0.2 setosa
13 4.8 3.0 1.4 0.1 setosa
14 4.3 3.0 1.1 0.1 setosa
15 5.8 4.0 1.2 0.2 setosa
16 5.7 4.4 1.5 0.4 setosa
17 5.4 3.9 1.3 0.4 setosa
18 5.1 3.5 1.4 0.3 setosa
19 5.7 3.8 1.7 0.3 setosa
20 5.1 3.8 1.5 0.3 setosa
21 5.4 3.4 1.7 0.2 setosa
22 5.1 3.7 1.5 0.4 setosa
23 4.6 3.6 1.0 0.2 setosa
24 5.1 3.3 1.7 0.5 setosa
25 4.8 3.4 1.9 0.2 setosa
26 5.0 3.0 1.6 0.2 setosa
27 5.0 3.4 1.6 0.4 setosa
28 5.2 3.5 1.5 0.2 setosa
29 5.2 3.4 1.4 0.2 setosa
30 4.7 3.2 1.6 0.2 setosa
31 4.8 3.1 1.6 0.2 setosa
32 5.4 3.4 1.5 0.4 setosa
33 5.2 4.1 1.5 0.1 setosa
34 5.5 4.2 1.4 0.2 setosa
35 4.9 3.1 1.5 0.2 setosa
36 5.0 3.2 1.2 0.2 setosa
37 5.5 3.5 1.3 0.2 setosa
38 4.9 3.6 1.4 0.1 setosa
39 4.4 3.0 1.3 0.2 setosa
40 5.1 3.4 1.5 0.2 setosa
41 5.0 3.5 1.3 0.3 setosa
42 4.5 2.3 1.3 0.3 setosa
43 4.4 3.2 1.3 0.2 setosa
44 5.0 3.5 1.6 0.6 setosa
45 5.1 3.8 1.9 0.4 setosa
46 4.8 3.0 1.4 0.3 setosa
47 5.1 3.8 1.6 0.2 setosa
48 4.6 3.2 1.4 0.2 setosa
49 5.3 3.7 1.5 0.2 setosa
50 5.0 3.3 1.4 0.2 setosa
# note that you can also just do this all on one line:
setosa <- iris[iris$Species == "setosa",]
Subsetting sepal length
Use the
>operator to create a new data frame that is the subset ofiriswith sepal length greater than 5.0.Solution
iris.new <- iris[iris$Sepal.Length > 5,]
Comparing variables – petal aspect ratio
Create a new data frame that includes all flowers for which the petal length is less than 3x the petal width.
Solution
iris.new <- iris[iris$Petal.Length < 3 * iris$Petal.Width,]
Combining concepts – subsetting and counting
How many flowers of each species in the current data set have petal length less than 3x the petal width?
Solution
# First subset on the aspect ratio as in the previous exercise iris.new <- iris[iris$Petal.Length < 3 * iris$Petal.Width,] # Now we have several options. # Option 1: Subset on species, then look at dimensions: iris.setosa <- iris.new[iris.new$Species == "setosa",] iris.versicolor <- iris.new[iris.new$Species == "versicolor",] iris.virginica <- iris.new[iris.new$Species == "virginica",] dim(iris.setosa)[1] # index dimensions to return # of rows[1] 1dim(iris.versicolor)[1][1] 8dim(iris.virginica) [1][1] 34# Option 2: Use logical == combined with sum() to count each # species name in the new data frame sum(iris.new$Species == "setosa")[1] 1sum(iris.new$Species == "versicolor")[1] 8sum(iris.new$Species == "virginica")[1] 34
Reading data from a tabulated file (.csv, .txt)
In the majority of cases, your data will be stored in some type of external file. The most common way that scientists store data is in Excel spreadsheets. While there are R packages designed to access data directly from Excel spreadsheets–the best I have found so far is readxl, for those who have need–these packages are often difficult and non-intuitive to use. Opening Excel spreadsheets from R is also slow, and it is not uncommon for large datasets to exceed the size limits of Excel. It is usually easier and faster to save tabular data in one or more comma-separated values (CSV or .csv) or text (TXT or .txt) files and then use R’s built in functionality to read and manipulate the data. Both file types are primitive forms of spreadsheet data anyway, in which data is stored as text with distinct data elements separated by a delimiting character–a comma , for .csv files and usually a tab (denoted \t in R) for .txt files.
Thankfully, Excel is capable of opening, editing, and saving both .csv and .txt files, so we can go back and forth between R and Excel when we find it convenient.
Here we will learn how to read external data from a .csv or .txt file, and write data modified or generated in R to a new .csv or .txt file. We will also explore the arguments that allow you read and write the data correctly for your needs.
Loading external data into a data frame
Let’s start by loading some sample data in .csv format using the read.csv() function and take a look at the first few rows of the contents using the head() function:
dat <- read.csv(file = 'data/sample.csv')
head(dat)
ID Gender Group BloodPressure Age Aneurisms_q1 Aneurisms_q2
1 Sub001 m Control 132 16.0 114 140
2 Sub002 m Treatment2 139 17.2 148 209
3 Sub003 m Treatment2 130 19.5 196 251
4 Sub004 f Treatment1 105 15.7 199 140
5 Sub005 m Treatment1 125 19.9 188 120
6 Sub006 M Treatment2 112 14.3 260 266
Aneurisms_q3 Aneurisms_q4
1 202 237
2 248 248
3 122 177
4 233 220
5 222 228
6 320 294
Open up the same file using a basic text editor (e.g. Notepad in Windows; TextEdit in MacOS). You should see something like this:
“ID”,”Gender”,”Group”,”BloodPressure”,”Age”,”Aneurisms_q1”,”Aneurisms_q2”,”Aneurisms_q3”,”Aneurisms_q4” “Sub001”,”m”,”Control”,132,16,114,140,202,237 “Sub002”,”m”,”Treatment2”,139,17.2,148,209,248,248 “Sub003”,”m”,”Treatment2”,130,19.5,196,251,122,177 “Sub004”,”f”,”Treatment1”,105,15.7,199,140,233,220 “Sub005”,”m”,”Treatment1”,125,19.9,188,120,222,228 “Sub006”,”M”,”Treatment2”,112,14.3,260,266,320,294
A couple of things to notice:
- The first line contains column headers.
- The
,delimiters tell theread.csv(...)function where the breaks in the data are located; that is, which data to put in which column of the output data frame. - Each new line is also a delimiter that tells
read.csv(...)which data to put in which row in the output data frame - In this case, all strings are contained within quotation marks
"". This is an optional way to tell the program where character strings start and stop.
On the topic of using quotation marks, open sample-noquotes.csv in your text editor to see the quote-free option:
ID,Gender,Group,BloodPressure,Age,Aneurisms_q1,Aneurisms_q2,Aneurisms_q3,Aneurisms_q4 Sub001,m,Control,132,16,114,140,202,237 Sub002,m,Treatment2,139,17.2,148,209,248,248 Sub003,m,Treatment2,130,19.5,196,251,122,177 Sub004,f,Treatment1,105,15.7,199,140,233,220 Sub005,m,Treatment1,125,19.9,188,120,222,228 Sub006,M,Treatment2,112,14.3,260,266,320,294
And note that the default read.csv(...) treats this file identically:
dat2 <- read.csv(file = 'data/sample-noquotes.csv')
head(dat2)
ID Gender Group BloodPressure Age Aneurisms_q1 Aneurisms_q2
1 Sub001 m Control 132 16.0 114 140
2 Sub002 m Treatment2 139 17.2 148 209
3 Sub003 m Treatment2 130 19.5 196 251
4 Sub004 f Treatment1 105 15.7 199 140
5 Sub005 m Treatment1 125 19.9 188 120
6 Sub006 M Treatment2 112 14.3 260 266
Aneurisms_q3 Aneurisms_q4
1 202 237
2 248 248
3 122 177
4 233 220
5 222 228
6 320 294
Whether to use quotes in your data files is up to you. The default behavior of read.csv(...) is smart enough to figure this out on it’s own. However, the read.csv(...) has an argument called quote that dictates how to interpret quotation marks.
Mis-loading quoted data
What if we force
read.csv()to ignore the quotes when they are present? Before you run any code, think about what will happen to the data in quotes if we tell R that there are no “quote” characters.dat3 <- read.csv(file = 'data/sample.csv', quote="") head(dat3)Solution
The quotes are now included as part of the character arguments. It also messed up the header formatting, appending an “X” to the beginning and surrounding text with “.”ss. Perhaps surprisingly, it does interpret teh numbers as numbers, rather than number “characters” with quotation marks around them.
What happens if we don’t assign the output of read.csv(...) to a variable?
read.csv(file = 'data/sample.csv')
As with any other function call, if the output is not explicitly assigned, it will be dumped into the console window. This can be annoying with large data files. Remember to assign your read...() functions!
Changing Delimiters
The default delimiter in the read.csv() function is a comma ,, but you can use essentially any set of characters as a delimiter. read.csv(...) is actually a special case of a more general function called read.table(...), with the delimiter argument (which is defined by the argument sep, for “separator”) set to , by default. Check out the help file for these functions:
?read.table # note that `?read.csv` brings up the same help document.
Under the Usage section there are multiple functions listed, including read.table(...) and read.csv(...). In the parentheses for each function there is a list of arguments. Since read.table(...) is the parent function, all arguments are listed. Only arguments with different default values (as indicated by the =) are listed for read.csv(...). A default value is the value that each argument assumes when you do not explicitly enter a value. For example, read.table(...) assumes that a data file has no header (header = FALSE) and no delimiting character (sep = "") while read.csv(...) assumes that a data file does have a header (header = TRUE) and a comma as the delimiting character (sep = ","), unless you specify otherwise.
If your data is stored in a tab-delimited text file, you will need to use read.table(...) with a different delimiting character, or another of the associated functions called read.delim(...) which has defaults to a tab-delimited file format. Note that to define a tab as a delimiting character, you have to use \t.
Let’s give it a try using a copy of the sample.csv data saved as a tab-delimited sample.txt file.
Note: From the help file, read.delim(...) defaults to header = TRUE while we have to explicitly define it when using read.table(...). We will talk about what this means in the next section.
# note that read
dat4 <- read.delim(file = 'data/sample.txt')
dat5 <- read.table(file = 'data/sample.txt', header = TRUE)
head(dat4)
ID Gender Group BloodPressure Age Aneurisms_q1 Aneurisms_q2
1 Sub001 m Control 132 16.0 114 140
2 Sub002 m Treatment2 139 17.2 148 209
3 Sub003 m Treatment2 130 19.5 196 251
4 Sub004 f Treatment1 105 15.7 199 140
5 Sub005 m Treatment1 125 19.9 188 120
6 Sub006 M Treatment2 112 14.3 260 266
Aneurisms_q3 Aneurisms_q4
1 202 237
2 248 248
3 122 177
4 233 220
5 222 228
6 320 294
head(dat5)
ID Gender Group BloodPressure Age Aneurisms_q1 Aneurisms_q2
1 Sub001 m Control 132 16.0 114 140
2 Sub002 m Treatment2 139 17.2 148 209
3 Sub003 m Treatment2 130 19.5 196 251
4 Sub004 f Treatment1 105 15.7 199 140
5 Sub005 m Treatment1 125 19.9 188 120
6 Sub006 M Treatment2 112 14.3 260 266
Aneurisms_q3 Aneurisms_q4
1 202 237
2 248 248
3 122 177
4 233 220
5 222 228
6 320 294
Now let’s take a closer look at a couple of useful arguments in read.table(...) family of functions.
The header argument
The default for read.csv(...) and read.delim(...) is to set the header argument to TRUE. This means that the first row of values in the .csv or .txt is used to define the column names for the output data frame. If your dataset does not have a header, set the header argument to FALSE.
Mis-loading data with headers
What happens if you forget to put
header = FALSEwhen loading a .csv file withread.csv()? The default value isheader = TRUE, which you can check with?read.csvorhelp(read.csv). What do you expect will happen if > you leave the default value?Before you run any code, think about what will happen to the first few rows of your data frame, and its overall size. Then run the following code and see if your expectations agree:
dat6 <- read.csv(file = 'data/sample.csv', header = FALSE) head(dat6)Solution
The
read.csv(...)function sets the column names as the default values (V1, V2, V3, ...) and treats the first row of thesample.csvfile as the first row of data. Clearly this is not the desired behavior for this data set, but it will be useful if you have a dataset without headers. Note that theVis used to start the column names, since column names must follow the usual variable naming rules, which would be violated with just a number.
The stringsAsFactors Argument
The stringsAsFactors argument tells R whether to treat imported data represented by a text string as a character or factor data type. Up until R version 4.0 was released, I introduced the stringsAsFactors argument as one of the most important in read.csv(...), particularly if you are working with categorical data. This is because the default behavior of R used to be to convert characters into factors. Because many relevant functions behave differently when confronted with a character vector than they do when confronted with a factor, this often resulted in unexpected behavior. In R 4.0 they updated the default behavior, and now strings of text are loaded as character data, rather than factors. This argument is still important, but the default behavior is now more intuitive and results in fewer frustrations.
The major problem is that, as previously discussed, factors do not let you add new data to a vector unless another element with that value us already present. As an example, let’s look at the car-speeds.csv data set. We find out that the data collector was color blind, and accidentally recorded green cars as being blue. In order to correct the data set, let’s replace ‘Blue’ with ‘Green’ in the $Color column:
# First - load the data with characters treated as factors and take a look at
# what information is available
carSpeeds <- read.csv(file = 'data/car-speeds.csv', stringsAsFactors = TRUE)
head(carSpeeds)
Color Speed State
1 Blue 32 NewMexico
2 Red 45 Arizona
3 Blue 35 Colorado
4 White 34 Arizona
5 Red 25 Arizona
6 Blue 41 Arizona
# Next use indexing to replace all `Blue` entries in the Color column with
# 'Green'
carSpeeds$Color[carSpeeds$Color == 'Blue'] <- 'Green'
Warning in `[<-.factor`(`*tmp*`, carSpeeds$Color == "Blue", value =
structure(c(NA, : invalid factor level, NA generated
head(carSpeeds, 10)
Color Speed State
1 <NA> 32 NewMexico
2 Red 45 Arizona
3 <NA> 35 Colorado
4 White 34 Arizona
5 Red 25 Arizona
6 <NA> 41 Arizona
7 <NA> 34 NewMexico
8 Black 29 Colorado
9 White 31 Arizona
10 Red 26 Colorado
What happened? Because we loaded the colors of the cars (represented by text strings) as factors, Green was not included in the original set of colors. Thus it was not included as a level in the factor. When we tried to replace Blue with Green, R changed each value to NA because Green was not included as a valid factor level.
To see the internal structure, we can use another function, str(). In this case, the data frame’s internal structure includes the format of each column, which is what we are interested in.
str(carSpeeds)
'data.frame': 100 obs. of 3 variables:
$ Color: Factor w/ 5 levels " Red","Black",..: NA 1 NA 5 4 NA NA 2 5 4 ...
$ Speed: int 32 45 35 34 25 41 34 29 31 26 ...
$ State: Factor w/ 4 levels "Arizona","Colorado",..: 3 1 2 1 1 1 3 2 1 2 ...
We can see that the $Color and $State columns are factors and $Speed is a numeric column.
One way to solve this problem would be to add the Green level to the Color factor, but there is a simpler way. Let’s reload the dataset using stringsAsFactors = FALSE, and see what happens when we try to replace ‘Blue’ with Green in the $Color column:
# First - load the data and take a look at what information is available
carSpeeds <- read.csv(file = 'data/car-speeds.csv', stringsAsFactors = FALSE)
str(carSpeeds)
'data.frame': 100 obs. of 3 variables:
$ Color: chr "Blue" " Red" "Blue" "White" ...
$ Speed: int 32 45 35 34 25 41 34 29 31 26 ...
$ State: chr "NewMexico" "Arizona" "Colorado" "Arizona" ...
# Next use indexing to replace all `Blue` entries in the Color column with 'Green'
carSpeeds$Color[carSpeeds$Color == 'Blue'] <- 'Green'
carSpeeds$Color
[1] "Green" " Red" "Green" "White" "Red" "Green" "Green" "Black" "White"
[10] "Red" "Red" "White" "Green" "Green" "Black" "Red" "Green" "Green"
[19] "White" "Green" "Green" "Green" "Red" "Green" "Red" "Red" "Red"
[28] "Red" "White" "Green" "Red" "White" "Black" "Red" "Black" "Black"
[37] "Green" "Red" "Black" "Red" "Black" "Black" "Red" "Red" "White"
[46] "Black" "Green" "Red" "Red" "Black" "Black" "Red" "White" "Red"
[55] "Green" "Green" "Black" "Green" "White" "Black" "Red" "Green" "Green"
[64] "White" "Black" "Red" "Red" "Black" "Green" "White" "Green" "Red"
[73] "White" "White" "Green" "Green" "Green" "Green" "Green" "White" "Black"
[82] "Green" "White" "Black" "Black" "Red" "Red" "White" "White" "White"
[91] "White" "Red" "Red" "Red" "White" "Black" "White" "Black" "Black"
[100] "White"
That’s better! And we can see how the data now is read as character instead of factor.
Note that there are certainly times when we do want text strings represented by factor variable types. Recall the example in last week’s On Your Own section dealing with ordered categorical data. Just be aware of the data types present in your data frames and how they impact the way you interact with your data.
Writing tabular data to a file (.csv, .txt)
After altering our cars dataset by replacing ‘Blue’ with ‘Green’ in the $Color column, we now want to save the output. The read.table(...) function family has a corresponding set of write.table(...) functions. These functions have a familiar set of arguments:
?read.table
Here is the basic format for writing a table:
# Export the data. The write.csv() function requires a minimum of two
# arguments, the data to be saved and the name of the output file.
write.csv(carSpeeds, file = 'data/car-speeds-corrected.csv')
If you open the file, you’ll see that it has header names, because the data had headers within R, but that there are also numbers in the first column.
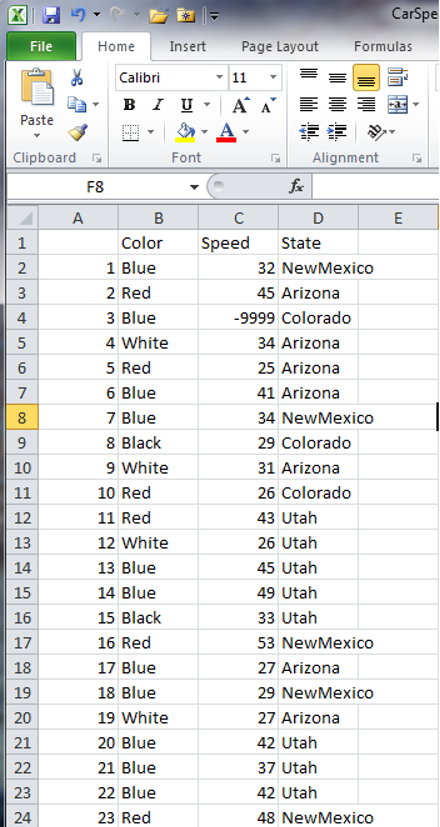
This extra column can cause problems when trying to open data in some other programs, and in most cases you won’t want to explicitly name the rows. This can be easily disabled with a simple argument:
The row.names Argument
This argument allows us to set the names of the rows in the output data file. R’s default for this argument is TRUE, and since it does not know what else to name the rows for the cars data set, it resorts to using row numbers. Unlike the recent change to the stringsAsFactors argument for reading data, this is an argument that defaults to something that is non-intuitive. To turn row names/numbers off, we can set row.names to FALSE:
write.csv(carSpeeds, file = 'data/car-speeds-cleaned.csv', row.names = FALSE)
Now we see:
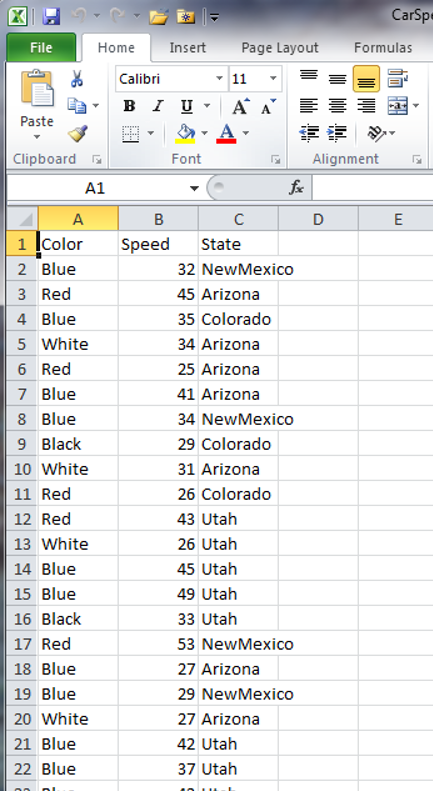
Setting column names
There is also a
col.namesargument, which can be used to set the column names for a data set without headers. If the data set already has headers (e.g. we used theheaders = TRUEargument when importing the data) then acol.namesargument will be ignored.
The na Argument
By default, R will export missing data in a dataset as NA. There are times when we want to specify certain values for missing data in our datasets (e.g. when we are going to pass the data to a program that only accepts -9999 or NaN as a “no data available” value). In this case, we want to set the NA value of our output file to the desired value, using the na argument. Let’s see how this works:
# First, replace the speed in the 3rd row with NA, by using an index (square
# brackets to indicate the position of the value we want to replace)
carSpeeds$Speed[3] <- NA
head(carSpeeds)
Color Speed State
1 Green 32 NewMexico
2 Red 45 Arizona
3 Green NA Colorado
4 White 34 Arizona
5 Red 25 Arizona
6 Green 41 Arizona
write.csv(carSpeeds, file = 'data/car-speeds-corrected-na.csv', row.names = FALSE)
Now we’ll set NA to -9999 when we write the new .csv file:
# Note - the na argument requires a string input
write.csv(carSpeeds,
file = 'data/car-speeds-corrected-9999.csv',
row.names = FALSE,
na = '-9999')
And we see:
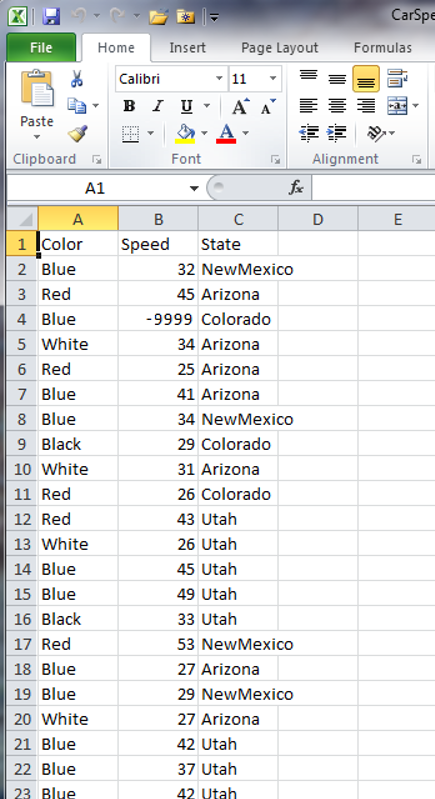
Key Points
All the indexing and subsetting that works on matrices also works on data frames.
Use
object[x, y]to select a single element from a data frame.Each column of a data frame can be directly addressed by specifying the column name using the
$operator (e.g.mydataframe$column1).Data in data structures can by accessed by specifying the appropriate index, by logical vector, or by column/row name (if defined).
Import data from a .csv or .txt file using the
read.table(...),read.csv(...), andread.delim(...)functions.Write data to a new .csv or .txt file using the
write.table(...)andwrite.csv(...)functions.